This post is also available in:
 עברית (Hebrew)
עברית (Hebrew)
Scientists have been using pandemic and epidemic simulation models for a long time to help determine how a large-scale disease will potentially spread, but those simulations can take anywhere from a couple of weeks to several months to complete. A recent study offers a new approach to epidemic modeling that could drastically speed up the process.
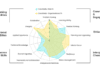
The study uses sparsification, a method from graph theory and computer science, to identify which links in a network are the most important for the spread of disease.
By focusing on critical links, the authors found they could reduce the computation time for simulating the spread of diseases through highly complex social networks by 90% or more.
“Epidemic simulations require substantial computational resources and time to run, which means your results might be outdated by the time you are ready to publish,” says lead author Alexander Mercier, a former Undergraduate Research Fellow at the Santa Fe Institute and now a Ph.D. student at the Harvard T.H. Chan School of Public Health.
“Our research could ultimately enable us to use more complex models and larger data sets while still acting on a reasonable timescale when simulating the spread of pandemics such as COVID-19.” Stated Mercier.
Prepared to dive into the world of futuristic technology? Attend INNOTECH 2023, the international convention and exhibition for cyber, HLS and innovation at Expo, Tel Aviv, on March 29th-30th
Interested in sponsoring / a display booth at the 2023 INNOTECH exhibition? Click here for details!